I went to Marti Anderson’s talk at UC BIDS yesterday where she introduced a generalization of mixed model ANOVA which uses a correction factor in the estimation which essentially corrects for how random your random effects are. She showed that if your random effects are actually close to fixed, then you gain statistical power by setting the correction factor close to 0. This was all good until after the talk, somebody asked a question to the effect of “well I thought the big difference between fixed and random effect models was that your data don’t have to be normal for fixed effects, so how does that come into play here?”
The answer: it doesn’t. Nowhere in her talk did Marti mention a normality assumption the data. P-values for the were computed using permutation tests, which make no assumptions about the underlying distribution of your data. All you need to use a permutation test is an invariance or symmetry assumption about your data under the null hypothesis: for example, if biological sex had no relation to height, then the distributions of male and female heights should be identical and we relabeled people male or female at random, the height distributions wouldn’t change.
People often confuse the assumptions needed to estimate parameters from their models and the assumptions to create confidence intervals and do hypothesis tests. The most salient example is linear regression. Even without making any assumptions, you can always fit a line to your data. That’s because ordinary least squares is just an estimation procedure: given covariates X and outcomes Y, all you need to do is solve a linear equation. Does that mean your estimate is good? Not necessarily, and you need some assumptions to ensure that it is. These are that your measurement errors are on average 0, they have the same variance (homoskedasticity), and they’re uncorrelated. Then your regression line will be unbiased and your coefficient estimates will be “best” in a statistical sense (this is the Gauss-Markov theorem). Of course, for this to work you also need more data points than parameters, no (near) duplicated covariates, and a linear relationship between Y and X, but hopefully you aren’t trying to use ordinary least squares if these don’t hold for your data!
Notice that there are no normality assumptions for the Gauss-Markov theorem to hold; you just need to assume a few things about the distribution of your Y data that can sort of be checked. Normal theory only comes into play if you want to use “nice” well-known distributions to construct confidence intervals. If you assume your errors are independent and identically distributed Gaussian mean-0 noise with common variance, then you can carry out F-tests for model selection and use the t-distribution to construct confidence intervals and to test whether your coefficients are significantly different from 0.
This is what usual statistical packages spit out at you when you run a linear regression, whether or not the normality assumptions hold. The estimates of the parameters might be ok regardless, but your confidence intervals and p-values might be too small. This is where permutation tests come in: you can still test your coefficients using a distribution-free test.
As an example, I’ve simulated data and done ordinary least squares in R, then compared the output of the lm function to the confidence intervals I construct by permutation tests.* I’m just doing a simple univariate case so it’s easy to visualize. The permutation test will test the null hypothesis that the regression coefficient is 0 by assuming there is no relationship between X and Y, so pairs are as if at random. I scramble the order of the Xs to eliminate any relationship between X and Y, then rerun the linear model. Doing this 1000 times gives an approximate distribution of regression coefficients if there were no association.
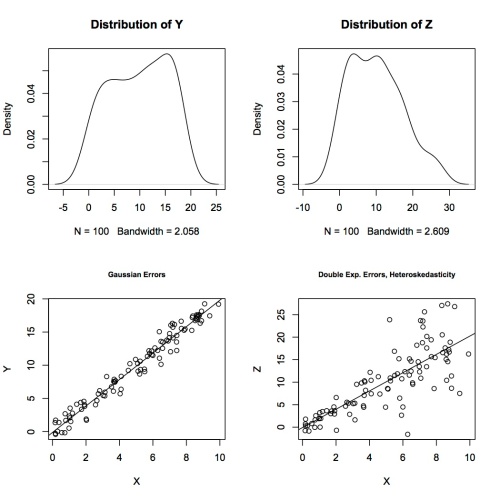
I generated 100 random X values, then simulated errors in two ways. First, I generated independent and identically distributed Gaussian errors e1 and let Y=2X + e1. I ran linear models and permutation tests for both to get the following 95% confidence intervals:
> confint(mod1,2) # t test
2.5 % 97.5 %
X 1.917913 2.068841
> print(conf_perm1) # permutation test
X X
1.598273 2.388481
When the normality assumptions are true, then we get smaller confidence intervals using the t-distribution than using permutation tests. That makes sense – we lose efficiency when we ignore information we know.
Then I generated random double exponential (heavy-tailed distribution) errors e2 that increase with X, so they break two assumptions for using t-tests for inference, and let Z = 2X + e2. The least squares fit is very good, judging by the scatterplot of Z against X – the line goes right through the middle of the data. There’s nothing wrong with the estimate of the coefficient, but the inference based on normal theory can’t be right.
> confint(mod2,2) # t test
2.5 % 97.5 %
X 1.716465 2.348086
> print(conf_perm2) # permutation test
X X
1.531020 2.533531
Again, we get smaller confidence intervals for the t-test, but these are simply wrong. They are artificially small because we made unfounded assumptions in using the t-test intervals; perhaps this points to one reason why so many research findings are false. It’s this very reason that it’s important to keep in mind what assumptions you’re making when you carry out statistical inference on estimates you obtain from your data – nonparametric tests may be the way to go.
* My simulation code is on Github.
